On the following pages, you can find information about current projects at the Department of Biophysics.
Our work particularly focuses on two main areas: molecular biophysics, in which basic research-oriented questions about the molecular functions and dynamics of proteins are investigated, and biospectroscopy, in which application-oriented questions, especially new approaches for (early) diagnosis of cancer and neurodegenerative diseases, are investigated.
The medical biophotonic works are particularly carried out within the framework of the Protein Research Unit Ruhr within Europe (PURE). From 2018 onwards, the research facility for molecular protein diagnostics (ProDi) will be available at the Bochum Healthcare Campus in which university and clinical researchers around PURE scientists will develop innovative methods for the early diagnosis of oncological and neurodegenerative diseases.
Many of the molecular biophysical studies are carried out within SFB 642.
If you are interested in more information, please contact us.
On the following pages you can inform yourself about the methodical approach in the field of molecular biophysics and our current work on the subject of recombinant proteins.
To study protein dynamics, physicists, chemists, biochemists and biologists work closely together at the Department of Biophysics.
In the first step, the proteins are recombinantly produced using molecular biological methods and purified for biophysical investigations. In the next step, the three-dimensional spatial structure of a protein is determined by means of X-ray structure analysis. The determination of the three-dimensional spatial structure is always an important milestone. It provides an image of the static ground state.
The protein dynamics are characterized by time-resolved spectroscopic methods. Thereby, the time-resolved FTIR spectroscopic techniques, which are developed further at the Department of Biophysics, allow to decipher molecular reaction mechanisms of proteins with atomic resolution. Different reactions can be analyzed simultaneously over different time scales, ranging from a few nanoseconds to several seconds, using a combination of step-scan FTIR and rapid-scan FTIR.
The methodical development of new time-resolved infrared spectroscopic methods for the analysis of the functions of proteins is an important goal at the Department of Biophysics. The experimental methods are supplemented by biomolecular simulations, molecular dynamics simulations and quantum chemical QM/MM simulations. Thereby, the spatial information coded in the IR spectra can finally be decoded.
In the end, there is not only a static image of the ground state but a movie of the dynamics of protein function and interaction with atomic resolution.
Based on the results of many publications, it was finally possible to produce movies of the light-driven proton pump bacteriorhodopsin and the GTPase mechanism of the Ras protein. The results are summarized in reviews.
At the Department of Biophysics, the investigated proteins are produced recombinantly. For this purpose, the genes coding for proteins are cloned and expressed. Essentially, the Escherichia coli system is used. For microbial cultivation, a number of shaking incubators and fermenters with volumes ranging from 1 to 20 liters of culture volume as well as the necessary infrastructure for downstream processing (cold rooms, centrifuges, devices for cell disruption) are available. For the biocompatible purification of proteins, various chromatography devices (2 Äkta-purifier and 3 Äkta-prime) are in operation.
Special expertise is provided for the isolation and analysis of integral membrane proteins, in particular light-, redox- and ATP-driven ion pumps. In addition to E. coli, proteins and their site specific variants are also produced in the archaeon Halobacterium salinarum (bacteriorhodopsin) and in the eukaryotic yeast Pichia pastoris (microbial retinal proteins).
Besides, a separate cell culture laboratory is used for the cultivation of insect and mammalian cells on a medium scale.
If you would like to find out more about the molecular biology work at the Department of Biophysics, please go on reading here.
(61) Rammelsberg, R., Huhn, G., Lübben, M., Gerwert, K.
Bacteriorhodopsin's Intramelecular Proton-Release Pathway consists of a Hydrogen-Bonded Network
Biochemistry 37, 5001-5009 (1998)
(63) Lübben, M., Prutsch, A., Mamat, B., Gerwert, K.
Elektron transfer induces side chain conformational changes of Glu-286 from cytochrome bo3
Biochemistry, 38, 2048-2056 (1999)
(127) Brucker, S., Gerwert, K., Kötting, C.
Tyr39 of Ran preserves the Ran·GTP gradient by inhibiting GTP hydrolysis
J. Mol. Biol. 401, 1–6 (2010)
(168e) Kuhne, J., Eisenhauer, K., Ritter, E., Hegemann, P., Gerwert, K., Bartl, F.
Early Formation of the Ion-Conducting Pore in Channelrhodopsin-2
Angew. Chem. Int. Ed. 2015, 54, 4953–4957
In order to determine the 3D structure of proteins, we mainly use X-ray crystallography. First, protein crystals are grown (for this, the samples have to be extremely homogenous), which are then measured on our X-ray diffractometer. Good crystals are additionally measured at synchrotron radiation sources (e.g. in Grenoble, Hamburg or Villigen) in order to achieve optimum data quality.
From these measured data, we can then determine the electron density distribution in the crystal and thus the position of all amino acids as well as the positions of bound water molecules or ligands. A particular challenge in this respect still is the crystallization of integral membrane proteins for which we employ special techniques, such as crystallization in lipidic cubic phases.
Within the scope of our collaborations with the Protein Crystallography Group of Prof. Eckhard Hofmann, we were thus able to elucidate the structural basis for the function of important membrane proteins. Especially the parallel investigation of functional protein variants with X-ray structure analysis and FTIR techniques gave us new insights into the bacterial reaction center (Hermes et al. 2006) or bacteriorhodopsin (Wolf et al. 2010).
If you would like to find out more about the work of the Protein Crystallography Group, please go on reading here.
(110) Hermes, S., Stachnik, J.M., Onidas, D., Remy, A., Hofmann, E., Gerwert, K.
Proton uptake in the reaction center mutant L210DN from Rhodobacter sphaeroides via protonated water molecules
Biochemistry (45) 13741-13748 (2006)
Supporting information
(128 deutsch) Wolf, S., Freier, E., Potschies, M., Hofmann, E., Gerwert, K.
Gerichteter Protonentransfer in Membranproteinen mittels protonierter proteingebundener Wassermoleküle: eine Protonendiode
Angew. Chem. 2010, 122, 7041-7046
The Department of Biophysics is equipped with several modern FTIR spectrometers of the company Bruker (80V, 66Vs), which have been optimized by self-made developments of our department for the respective application. The evacuable beam paths of the spectrometers allow for rapid-scan (Gerwert et al. 1988, Gerwert et al. 1990) and step-scan measurements (Rammelsberg et al. 1997, patent: WO1999040413 A1). An experimental setup is shown in the left figure. With this, simultaneous reactions within one experiment can be observed from the nanosecond to the second time range.
The IR absorption changes of channelrhodopsin from 100 ns to 140 s are shown in the right figure. The reactions are initiated by light flashes of powerful excimer or dye lasers. Chromoproteins can be excited directly. Apart from that, photolabile triggers, photocaged compounds such as caged GTP, are available. The approach of time-resolved FTIR spectroscopy has been established for bacteriorhodopsin. The IR absorption changes as well as the associated kinetics of corresponding marker bands and the proton transfer path of the protein are shown simultaneously in the following movie.
Patent:
Gerwert, K.
"Method for time-resolved measurement of energy spectra of molecular states and device for carrying out said method"
WO1999040413 A1
(13) Gerwert, K.
Intramolekulare Proteindynamik untersucht mit zeitaufgelöster Fourier Transform Infrarot-Differenzspektroskopie
Ber. Bunsenges. Phys. Chem. 92, 978-982 (1988).
(22) Gerwert, K., Souvignier, G., Hess, B.
Simultaneous monitoring of light-induced changes in protein side-group protonation, chromophore, isomerization, and backbone motion of bacteriorhodopsin by time-resolved Fourier-transform infrared spectroscopy.
Proc. Natl. Acad. Sci. USA, 87, 9774-9778 (1990).
(55) Rink, Th., Riesle, J.,Oesterhelt, D., Gerwert, K., Steinhoff, H.-J.
Spin labeling studies of the conformational changes in the vicinity of D36,D38,T46 and E161 of bacteriorhodopsin during the photocycle
Biophysical Journal 73, 983-993 (1997)
ATR-FTIR spectroscopy is a surface-based technique that allows label-free and time-resolved analysis of protein-ligand interactions in the same way as done with the widely used SPR (surface plasmon resonance) spectroscopy. Because of the spectral resolution of infrared spectroscopy additionally the complete molecular information of protein-ligand interaction at every time point can be obtained (Kötting et al. 2012).
In the ATR technique, the infrared beam reflects within a high refractive index medium namely the internal reflection element (IRE). At the interface, a so-called evanescent field is formed which extends into the surrounding medium approximately 1 µm and interacts with the protein immobilized on the IRE. The ATR technology thus allows the measurement of liquids in a flow system in the infrared spectral range.
By forming membrane monolayers of natural lipids on the IRE and the in situ attachment of lipid-anchored Ras, this peripheral membrane protein could be investigated by infrared spectroscopy in a more native environment. It was shown that membrane-bound N-Ras has an upright molecular orientation. Simultaneous FRET studies have shown that N-Ras is membrane-bound as a dimer. This was a cover story in the Biophysical Journal (Güldenhaupt et al. 2012). Protein-protein interactions have also been investigated with the ATR system. For example, the membrane extraction of the GTPase Rab by a GDI was detected (Gavriljuk et al. 2013).
The freely accessible ATR surface allows targeted manipulation of the immobilized proteins, for example by the addition of ligands. These monolayers, which contain only a few µg of protein, can be used, e.g. by using multiple reflections, to detect very small conformational changes of the immobilized proteins and to investigate molecular reaction mechanisms (Pinkerneil et al. 2012, cover story). At the same time, the intrinsic polarization of the evanescent field is used to perform orientation analyses of monolayers of immobilized proteins and lipids.
The specific immobilization of proteins on the IRE is achieved in our department through different surface chemical techniques, which always work as a layer by layer modification of the ATR surface under simultaneous online spectral control. Thereby the first layer is a monolayer of thiols or silanes (SAM, self assembled monolayer), which form a covalent bond to the previously activated germanium surface of the ATR crystal (Schartner et al. 2013). The second layer is the protein interaction layer, which is coupled in the measurement cell to the SAM by means of NHS, maleimido or click chemistry. The protein interaction layer can consist of NTA groups, streptavidin or antibodies so that any HisTag, StrepTag or biotin-bearing proteins, or any protein compatible with the antibody used, can be immobilized.
The resulting surface concentration of the immobilized protein allows for time-resolved detection of ligand induced conformational changes in the time domain 10 s - 10 h (K4DD).
The antibody immobilization additionally allows certain disease-relevant marker proteins to be captured highly specific from complex liquids (e.g. blood plasma). On this basis we developed an immuno-ATR biosensor for diagnosis of Alzheimer's disease.
(136) Kötting, C., Güldenhaupt, J., Gerwert, K.
Time-resolved FTIR spectroscopy for monitoring protein dynamics exemplified by functional studies of Ras protein bound to a lipid bilayer
Chemical Physics, 396, 72-83 (2012)
(140) Pinkerneil, P., Güldenhaupt, J., Gerwert, K., Kötting, C.
Surface attached polyhistidine-tag proteins characterized by FTIR difference spectroscopy
ChemPhysChem 2012, 13, 2649 – 2653
(144) Güldenhaupt, J., Rudack, T., Bachler, P., Mann, D., Triola, G., Waldmann, H., Kötting, C. and Gerwert, K.
N-Ras Forms Dimers at POPC Membranes
Biophys. J. 103, 1585-1593 (2012)
Supporting Material
(149) Schartner, J., Güldenhaupt, J., Mei, B., Rögner, M., Muhler, M., Gerwert, K., Kötting, C.
Universal Method for Protein Immobilization on Chemically Functionalized Germanium Investigated by ATR-FTIR Difference Spectroscopy
J. Am. Chem. Soc. 135, 4079-4087 (2013)
(155) Gavriljuk, K., Itzen, A., Goody, R. S., Gerwert, K., Kötting, C.
Membrane extraction of Rab proteins by GDP dissociation inhibitor characterized using attenuated total reflection infrared spectroscopy
Proc. Natl. Acad. Sci., 110, 13380-13385 (2013)
Supporting Material
(174) El-Mashtoly, S.F., Yosef, H.K., Petersen, D., Mavarani, L., Maghnouj, A., Hahn, S.A., Kötting, C., Gerwert, K.
Label-free Raman spectroscopic imaging monitors the integral physiologically relevant drug responses in cancer cells
Anal. Chem. 87 (2015) 7297-7304
Supporting Material
At the Department of Biophysics, besides experimental methods, also computer-aided biomolecular simulations of macromolecules and proteins are performed. These include mainly molecular dynamics (MD-) simulations, that describe atoms via classical mechanics, and quantum mechanical (QM-) simulations, that also consider electron distributions. For this purpose we are equipped with an own high performance computer cluster (hpcpure - 1400 CPUs) and several GPU-powered systems that enable calculation of protein and macromolecule behavior with high spatio-temporal resolution (subatomic resolution with femtosecond time steps) over large periods of time (µs).
Calculation of theoretical IR spectra enables strong cooperation with the experiments that are performed at the department. On the one hand this enables verification and benchmarking of the simulation data and on the other hand gives high resolved models for the elucidation of experimental findings. This was shown e.g. for the calculation of FTIR spectra of small GTPases (Rudack et al. 2012), that enabled a detailed investigation of the hydrolysis mechanism in Ras proteins (Rudack et al. 2012), investigations of how single hydrogen bonds influence the nucleotide exchange in heterotrimeric G proteins (Schröter et al. 2015), simulations that elucidate the transport mechanism of ions in channelrhodopsin (Eisenhauer et al. 2012), and elucidation of the macromolecular organization of Ras dimers on membranes (Güldenhaupt et al. 2012).
(138) Eisenhauer K., Kuhne J., Ritter E., Berndt, A., Wolf S., Freier E., Bartl F., Hegemann P., Gerwert K.
In channelrhodopsin-2 E90 is crucial for ion selectivity and is deprotonated during the photocycle
Journal of Biological Chemistry, 287 (9), 6904-6911 (2012)
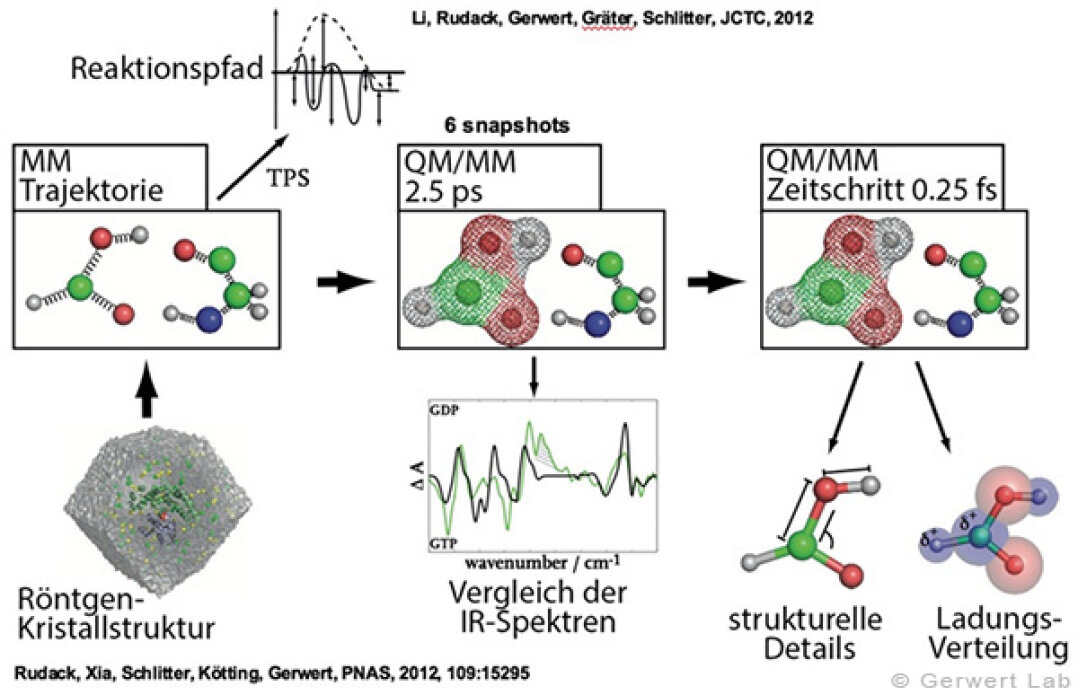
(142) Rudack, T., Fei X., Schlitter, J., Kötting, C., Gerwert, K.
Ras and GTPase-activating protein (GAP) drive GTP into a precatalytic state as revealed by combining FTIR and biomolecular simulations
Proc. Natl. Acad. Sci., 109, 15295-15300 (2012)
(143) Rudack, T., Fei X., Schlitter, J., Kötting, C., Gerwert, K.
The role of magnesium for geometry and charge in GTP hydrolysis, revealed by QM/MM simulations
Biophysical Journal, July 2012 (103), 293-302
Supporting Material
(144) Güldenhaupt, J., Rudack, T., Bachler, P., Mann, D., Triola, G., Waldmann, H., Kötting, C. and Gerwert, K.
N-Ras Forms Dimers at POPC Membranes
Biophys. J. 103, 1585-1593 (2012)
Supporting Material
(172) Schröter, G., Mann, D., Kötting, C., Gerwert, K.
Integration of Fourier Transform Infrared Spectroscopy, Fluorescence Spectroscopy, Steady-State Kinetics and Molecular Dynamics Simulations of Gαi1 distinguishes between the GTP Hydrolysis and GDP Release Mechanism
J. Biol. Chem. (2015) 290 (28) 17085-17095

By means of methods described in the part methodical approach, we investigate the reaction mechanisms and interactions of recombinant membrane proteins (bacteriorhodopsin, channelrhodopsin, photosynthetic reaction center) as well as the ones of small and heterotrimeric GTPases, which are bound to lipid membranes by membrane anchors.
Bacteriorhodopsin is a light-driven proton pump that has been intensively studied by research groups all over the world. Therefore it is nowadays considered as an important biophysical model system. Various new biophysical methods, for instance time-resolved FTIR difference spectroscopy (Gerwert 1988) have been developed in order to monitor the mechanism of this protein. Nowadays bR is used to inhibit neuronal processes in optogenetics.
As documented in various publications, we resolved the proton transfer reactions in great detail. The chromophore retinal is covalently bound to the protein via a protonated Schiff Base which is the central proton binding site of the protein. Upon light-induced all-trans- to 13-cis isomerization of the retinal the pK-value of the binding site is decreased. This leads to proton transfer from the Schiff Base to the nearby Asp85 in the L to M transition of the photocycle (Gerwert 1988). For the first time, we could detect this reaction using time-resolved FTIR difference spectroscopy (Gerwert et al. 1989; Gerwert et al. 1990).
The protonation of Asp85 causes a downward movement of Arg82 (Gerwert et al. 1989) as we predicted using pK-value calculations (Bashford, Gerwert 1992) and verified experimentally (Wolf et al. 2010). We identified a protonated water cluster that works as the proton release group of bR. It is the first time that such a structure has been detected in a protein (Garczarek, Gerwert 2006). In contrast to the dynamic protonated water clusters of pure water that were identified by nobel laureate Manfred Eigen the surrounding amino acids, especially Glu198, Glu204 und Arg82 play an important role for stabilizing charges (Wolf et al. 2014).
Asp96 is important for resetting the pump. In the M to N transition (Bashford, Gerwert 1992) Asp96 deprotonates which leads to reprotonation of the Schiff Base. We could demonstrate that for the first time (Gerwert et al. 1990). Thereby we identified the central reactions of the pump, which are the proton release to Asp85 and the proton uptake from Asp96 (Gerwert et al. 1989; Gerwert et al. 1990). Based on these results a spatially and temporally resolved proton pump model has been proposed which has now found its way into the textbooks (Gerwert 1992).
Three water molecules are arranged by a specific conformational change of the protein to a linear transient Grotthuß chain to allow long range proton transfer from Asp96 to the Schiff Base. For the first time we were able to reveal such a transient Grotthuß chain in a protein (Freier et al. 2011). Asp96 is reprotonated from the cytoplasmic side (Garczarek, Gerwert 2006). After re-isomerization of the retinal in the all-trans conformation Asp85 deprotonates in the O to bR transition.
Thereby the proton release group is reprotonated in this last step of the photocycle (Gerwert et al. 1990). This functional role of water molecules that we could identify for the first time is summarized in a review article (Gerwert et al. 2013).
Structure and proton transfer steps of bacteriorhodopsin
according to: Garczarek, F., Gerwert, K. Nature (439) 109-112 (2006)
bR - The Movie
according to: Gerwert, K., Freier, E., Wolf, S., Biochim. Biophys. Acta 1837 (2014) 606–613
QM/MM Simulations of Protein Bound Water Molecules
according to: Gerwert, K., Freier, E., Wolf, S., Biochim. Biophys. Acta 1837 (2014) 606–613
Protonated Water Cluster: Top View
according to: Garczarek, F., Gerwert, K.Nature (439) 109-112 (2006)
Transient (1ms) Linear Proton Conducting Water Chain
according to: Freier, E., Wolf, S., Gerwert, K. PNAS 108 (28), 11435-11439 (2011)
QM/MM Simulation of the Transient Water Chain
according to: Wolf, S., Freier, E., Cui, Q., Gerwert, K., J Chem Phys 2014, 141(22), 22D524
Channelrhodopsin is a light-activated ion channel that is used in optogenetics to investigate neuronal processes with high spatial and temporal resolution.
Like bacteriorhodopsin, channelrhodopsin is a microbial rhodopsin. Using the same methodical approach we established for bacteriorhodopsin, we could resolve the early opening of the channel both temporal and spatial together with Peter Hegemann (Eisenhauer et al. 2012, Kuhne et al. 2015). Especially the combination of step-scan FTIR spectroscopy, homology modelling and simulation of the dynamics of protein bound water molecules turned out to be the key to success. The essential difference of channelrhodopsin compared to the proton pump Bacteriorhodopsin is helix 2 with its numerous glutamates, especially Glu90.
We were able to show that the isomerization of the retinal leads to a destabilization of Glu90 which then flips downward and deprotonates (figure a). This opens the pore (figure b), water enters the hydrophobic vestibule above the pore and presses helix 2 outward so that the channel opens (lower figure).
This so called EHT-model can explain the behavior of different site specific mutants and provides the basis for the design of optimally adapted optogenetic tools. This work was the cover story of "Angewandte Chemie" in 2015.
(138) Eisenhauer K., Kuhne J., Ritter E., Berndt, A., Wolf S., Freier E., Bartl F., Hegemann P., Gerwert K.
In channelrhodopsin-2 E90 is crucial for ion selectivity and is deprotonated during the photocycle
Journal of Biological Chemistry, 287 (9), 6904-6911 (2012)
(168g) Kuhne, J., Eisenhauer, K., Ritter, E., Hegemann, P., Gerwert, K., Bartl, F.
Die frühe Entstehung der ionenleitenden Pore in Channelrhodopsin-2
Angew. Chem. 2015, 127, 5037-5041
Small GTPases are molecular switches that regulate numerous processes in cells. The most intensively investigated one is the Ras protein, which switches the signal for cell growth towards the nucleus. Oncogenic point mutations of Ras in the GTP binding site prevent the GTP hydrolysis from GTP to GDP. By this hydrolysis reaction, the GTPase is usually switched off and the growth signal transduction is interrupted. The figure above shows the gamma-phosphate attacking water molecule. This reaction is very slow in solution (200 days at 25 °C) but is accelerated in Ras by 5 orders of magnitude (20 min). GTPase-activating proteins (GAPs) accelerate by 5 additional orders of magnitude (100 ms) and thus the cell growth is controlled. For oncogenic mutations, this catalysis is inhibited and an uncontrolled permanent growth signal is generated. This contributes to cancer development.
We were able to elucidate the catalysis mechanism in detail by the combination of experimental and theoretical infrared spectroscopy: for the first time, we were able to trace the movement of the most important amino acid of GAPs, the arginine finger, into the catalytic binding pocket in a time-resolved manner and show that this process is entropy-driven (Kötting et al. 2008). The movement of the arginine finger induces a charge shift towards the product state (Rudack et al. 2012). Furthermore, GTP is stretched towards the direction of the transition state by protein binding, similar to a steel spring in a toy car. This facilitates the bond breakage. The results are summarized in the movie below.
With this approach, we have investigated further small GTPases. Eukaryotic cells contain 100-150 different small GTPases. Common to all GTPases is the G domain. Differences arise with the dynamics of the construction of the catalytic center consisting of the G domain and the complementary GAP. For example, the Ras-GAP (Kötting et al. 2006) and Rap-GAP complexes (Chakrabarti et al. 2007) form an intermediate during the GTP hydrolysis, in which the bond breakage has already been achieved, but the phosphate is still stably bound within the protein complex. Thus, the release of the phosphate from the protein is the rate limiting step. Our characterization of the intermediate in 2006 was the title story in PNAS (Kötting et al. 2006).
In contrast, the GTPases RhoA, Ran (Brucker et al. 2010) and Rab1 (Gavriljuk et al. 2012) with their respective GAPs show that the bond breakage is rate limiting. Differences also arise in the so-called arginine finger, which is catalytically decisive for most GTPases (Ras, Rho, Rab), but is not present in others at all (Ran). Common to all GTPases is an acid amide, which in some cases is an intrinsic amino acid of the GTPase, but in others is provided by a GAP. This was especially the case for Rab (Gavriljuk et al. 2012). The dynamic structure of the catalytic center determines the high specificity between the respective small GTPase and the associated GAP.
For the biological function of most GTPases, binding to the membrane is important, to which they are bound via lipid anchors. One differentiates, for example, between H-, N- and K-Ras. K-Ras plays a central role in cancer development. For the first time, we demonstrated that Ras is dimerized on membranes (Güldenhaupt et al. 2012). This mechanism now allows a completely new attack point for drugs. We have summarized our recent work on small GTPases in a review article (Kötting, Gerwert 2015). The dimerization of Ras, the influence on the growth signal transduction and the intervention of dimerization with small potential drugs is currently under intensive investigation in our laboratory. The nucleophilic attack of the water molecule and the development of potential drugs are also investigated.
(108) Kötting, C., Blessenohl, M., Suveyzdis, Y., Goody, R.S., Wittinghofer, A., Gerwert, K.
A phosphoryl transfer intermediate in the GTPase reaction of Ras in complex with its GTPase-activating protein
Proc. Natl. Acad. Sci USA (103) 13911-13916 (2006)
(111) Chakrabarti, P., Daumke, O., Suveyzdis, Y., Kötting, C., Gerwert, K., Wittinghofer, A.
Insight into catalysis of a unique GTPase reaction by a combined biochemical and FTIR approach
J. Mol. Biol. (367) 383-995 (2007)
(117) Kötting, C., Kallenbach, A., Suveyzdis, Y., Wittinghofer, A. Gerwert, K.
The GAP arginine finger movement into the catalytic site of Ras increases the activation entropy
Proc. Natl. Acad. Sci, 105, 6260-6265 (2008)
(127) Brucker, S., Gerwert, K., Kötting, C.
Tyr39 of Ran preserves the Ran·GTP gradient by inhibiting GTP hydrolysis
J. Mol. Biol. 401, 1–6 (2010)
(143) Rudack, T., Fei X., Schlitter, J., Kötting, C., Gerwert, K.
The role of magnesium for geometry and charge in GTP hydrolysis, revealed by QM/MM simulations
Biophysical Journal, July 2012 (103), 293-302
Supporting Material
(144) Güldenhaupt, J., Rudack, T., Bachler, P., Mann, D., Triola, G., Waldmann, H., Kötting, C. and Gerwert, K.
N-Ras Forms Dimers at POPC Membranes
Biophys. J. 103, 1585-1593 (2012)
Supporting Material
(147)Gavriljuk, K., Gazdag, E.-M., Itzen, A., Kötting, C., Goody, R.S., Gerwert, K.
Catalytic mechanism of a mammalian Rab·RabGAP complex in atomic detail
Proc. Natl. Acad. Sci., 109, 21348-21353 (2012)
(169) Kötting, C., Gerwert, K.
Review: What vibrations tell us about GTPases
Biol. Chem. 2015; 396(2): 131–144
Interaction network of the Ras protein
according to: Henrik te Heesen
Heterotrimeric GTPases have a switch mechanism very similar to the one of small GTPases. In particular, we study the deactivation of the Gα subunits, which is crucial for the regulation of numerous signal transduction pathways. Malfunctions of Gα proteins are the basis for many diseases such as fibrosis, cancer, whooping cough, and cholera. Here, the most important method is the time-resolved FTIR spectroscopy which can be used to follow the amino acids involved in the reaction, such as the intrinsic arginine finger and the catalytic glutamine, to determine their exact function. Using the same technique, we also study the mechanism of toxins that modify Gα proteins. The combination of the experimental results with biomolecular simulations allows the identification of further details of the reaction mechanism with highest resolution.
(101) Kötting, C., Gerwert, K.
Proteins in action monitored by Time-resolved FTIR Spectroscopy
Chem Phys. Chem (6) 1-8 (2005)
(169) Kötting, C., Gerwert, K.
Review: What vibrations tell us about GTPases
Biol. Chem. 2015; 396(2): 131–144
(172) Schröter, G., Mann, D., Kötting, C., Gerwert, K.
Integration of Fourier Transform Infrared Spectroscopy, Fluorescence Spectroscopy, Steady-State Kinetics and Molecular Dynamics Simulations of Gαi1 distinguishes between the GTP Hydrolysis and GDP Release Mechanism
J. Biol. Chem. (2015) 290 (28) 17085-17095